GitHub - c-guzman/wishbone: Tool to investigate, analyze, and compare TSS-MPRA data
4.6
$ 8.99
In stock
(218)
Product Description
GitHub - JSuryatenggara/ChIP-AP: An Integrated Analysis Pipeline for Unbiased ChIP seq Analysis

Security Threat Tool Design in Percona Monitoring and Management
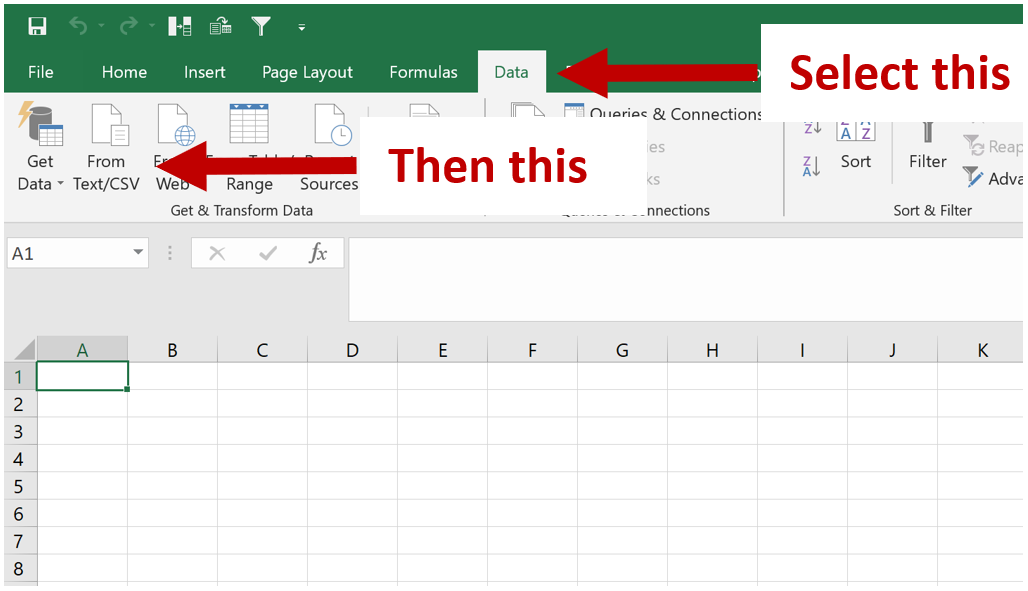
GitHub - JSuryatenggara/ChIP-AP: An Integrated Analysis Pipeline for Unbiased ChIP seq Analysis
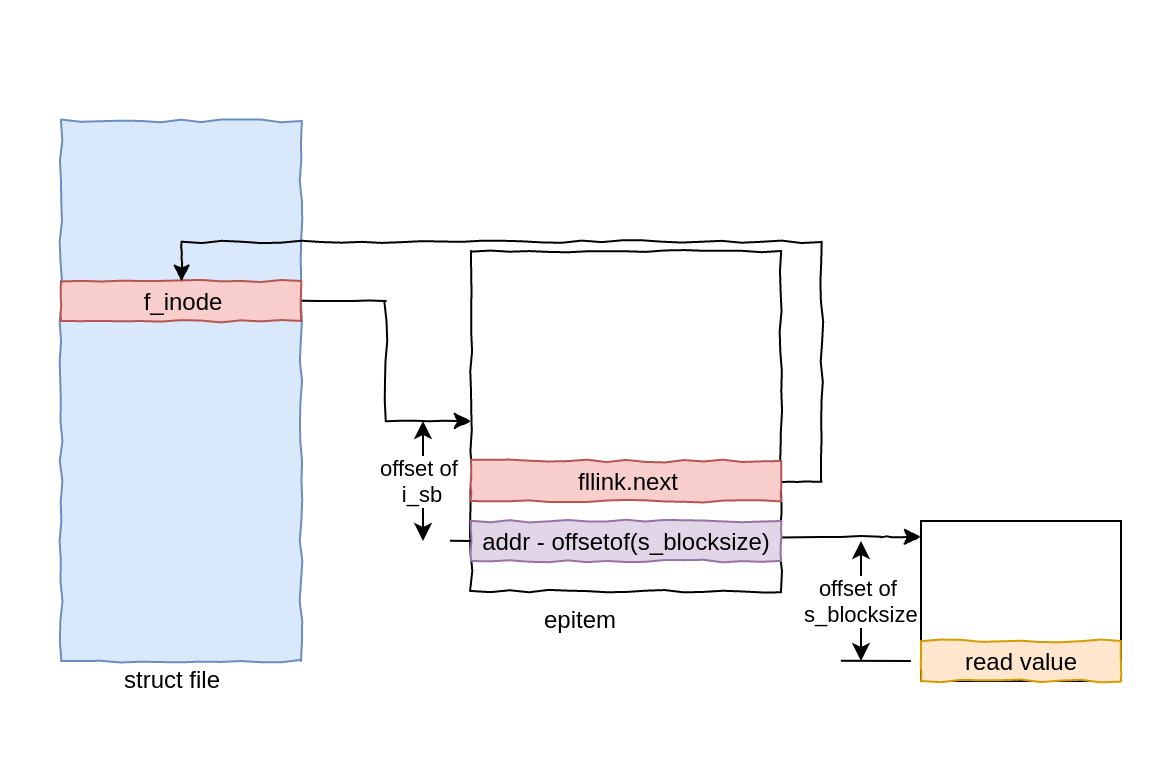
Exploiting CVE-2020-0041 - Part 2: Escalating to root
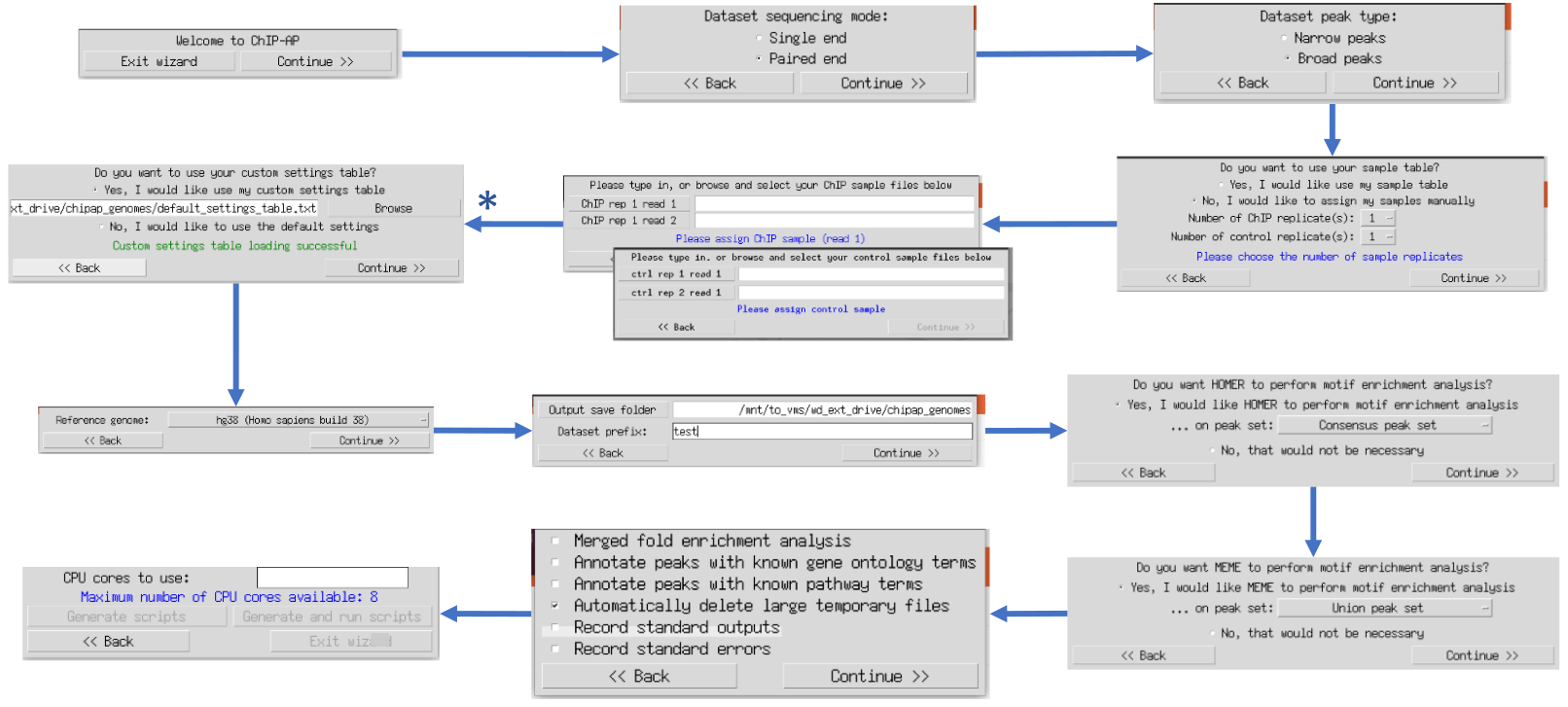
GitHub - JSuryatenggara/ChIP-AP: An Integrated Analysis Pipeline for Unbiased ChIP seq Analysis

PDF) Combining TSS-MPRA and sensitive TSS profile dissimilarity scoring to study the sequence determinants of transcription initiation
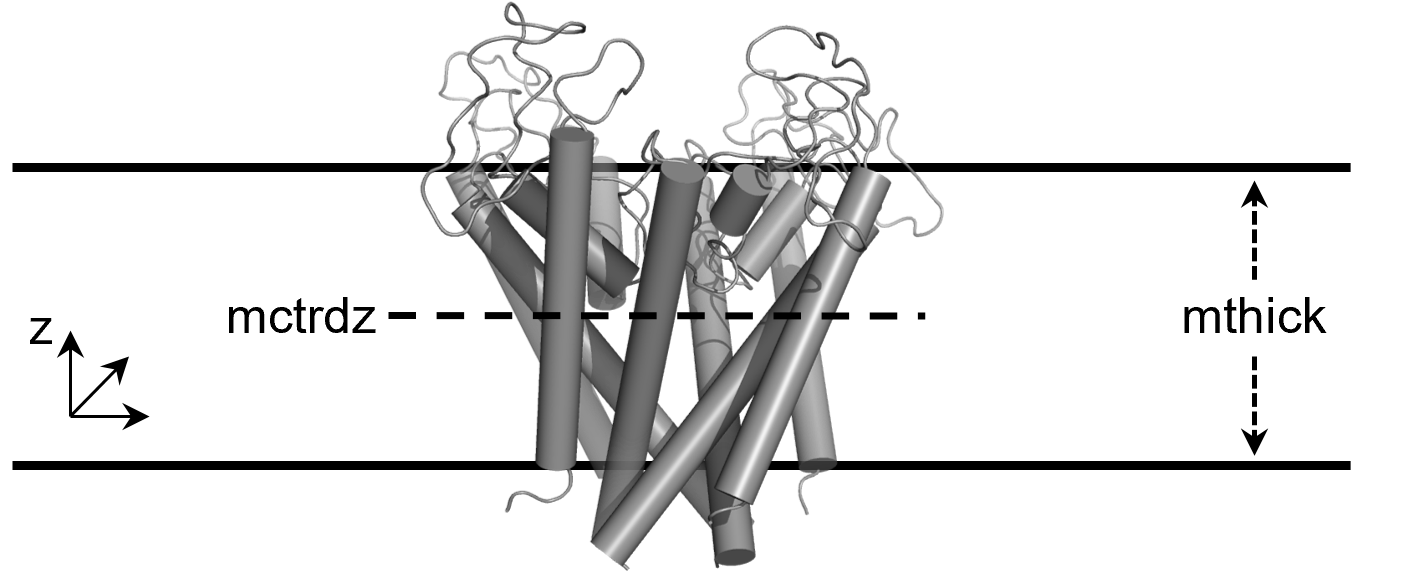
The input file - gmx_MMPBSA Documentation
GitHub - JSuryatenggara/ChIP-AP: An Integrated Analysis Pipeline for Unbiased ChIP seq Analysis

How to start contributing to Open Source.
GitHub - c-guzman/wishbone: Tool to investigate, analyze, and compare TSS-MPRA data

Interpretation of the unassigned junction table · Issue #170 · barricklab/breseq · GitHub
GitHub - JSuryatenggara/ChIP-AP: An Integrated Analysis Pipeline for Unbiased ChIP seq Analysis
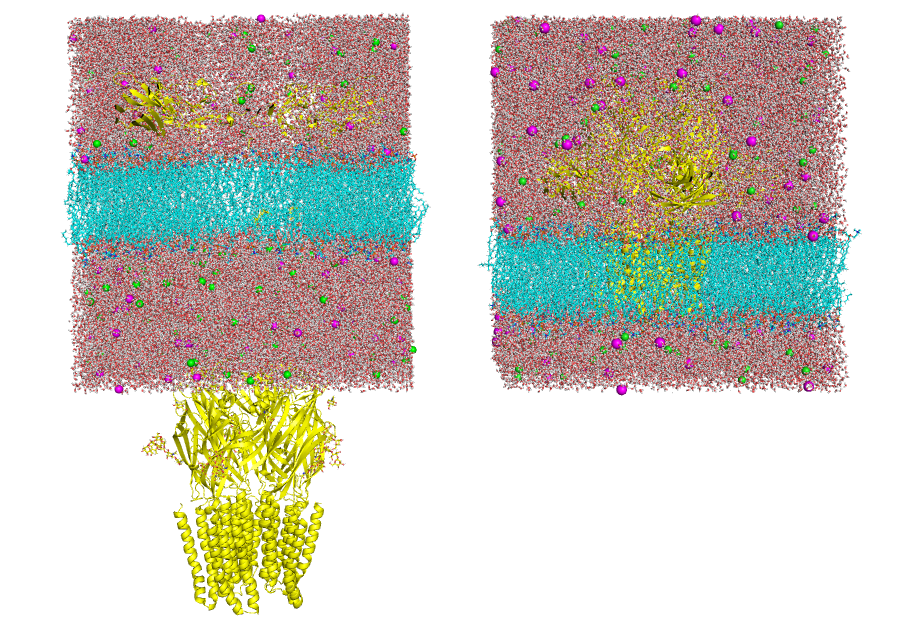
Running gmx_MMPBSA - gmx_MMPBSA Documentation

Use vc_nonSyn customized variant classifications plotmafSummary can not be displayed · Issue #748 · PoisonAlien/maftools · GitHub
GitHub - pereiramemo/AGS-and-ACN-tools: Fast and accurate Average Genome Size and 16S rRNA gene Average Copy Number computation in metagenomic data









